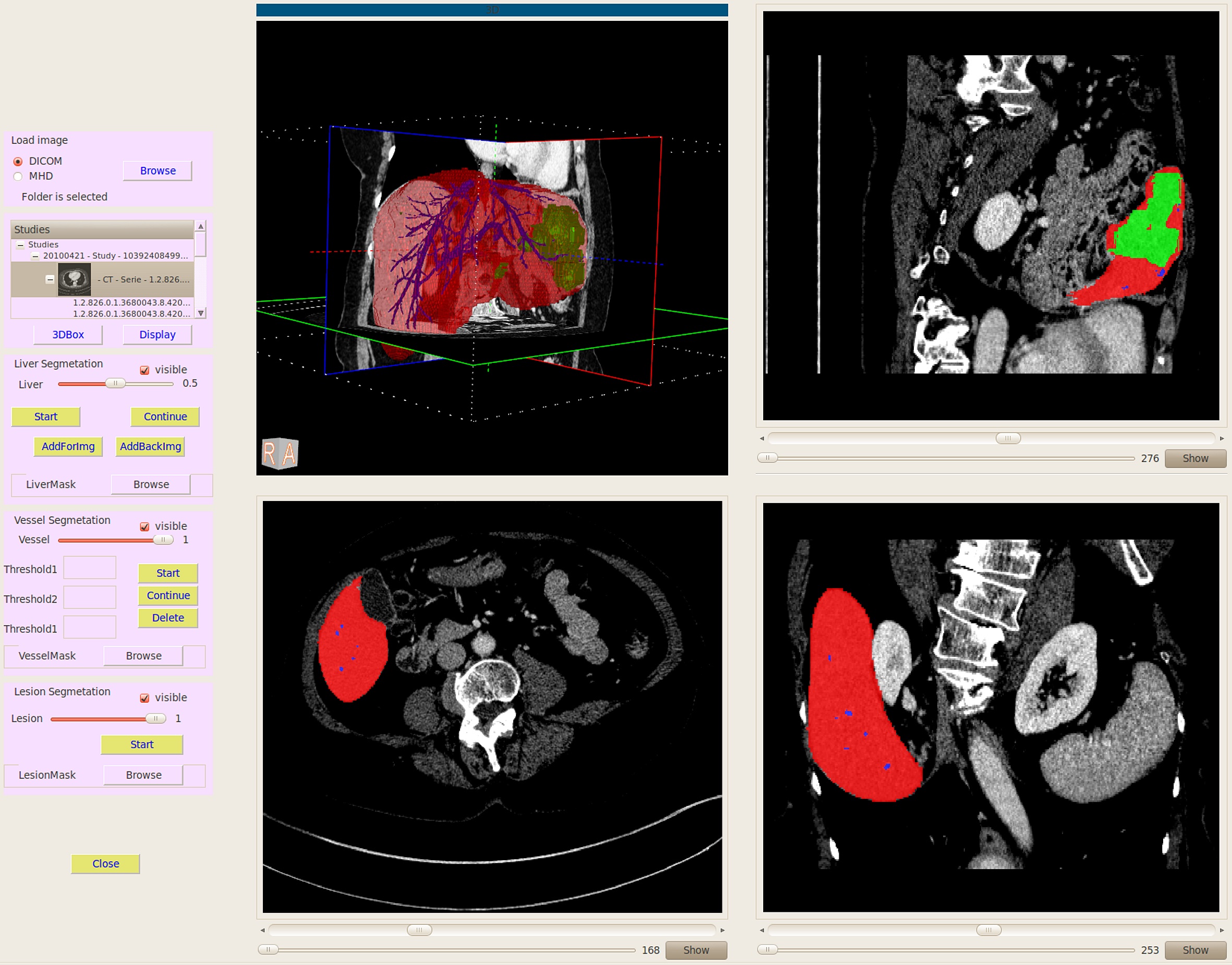
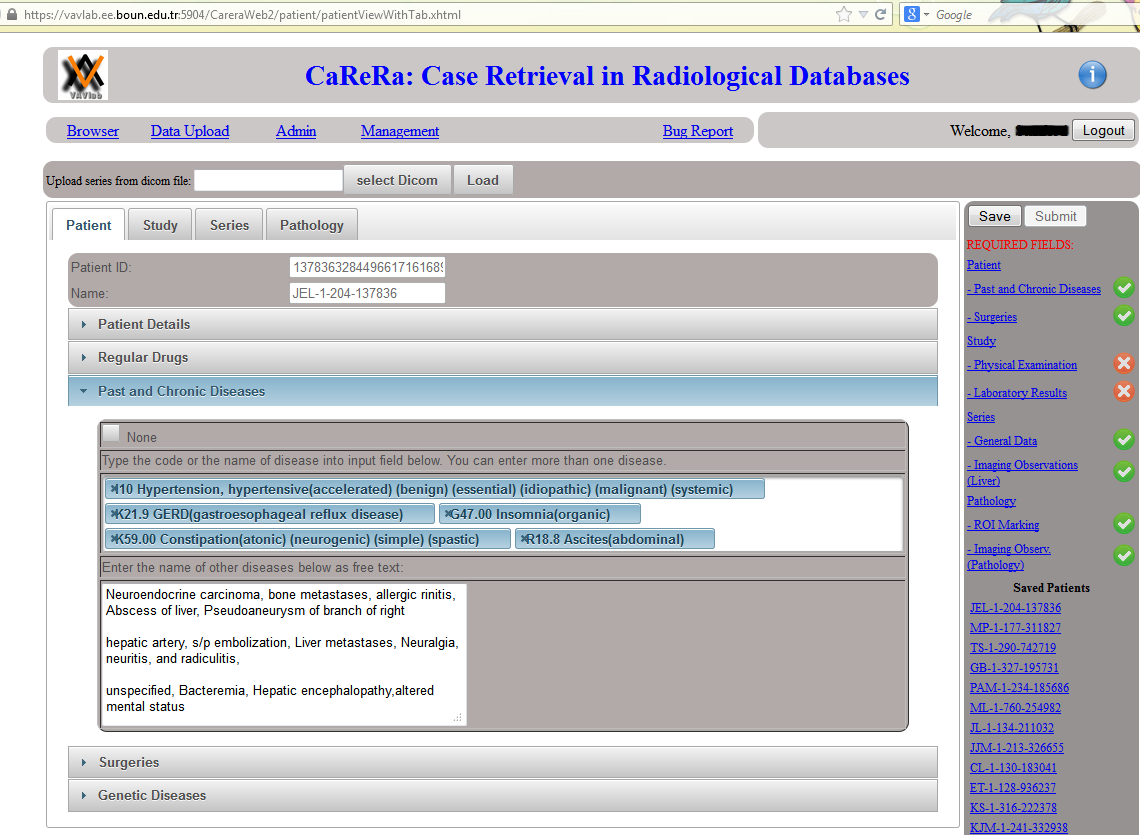
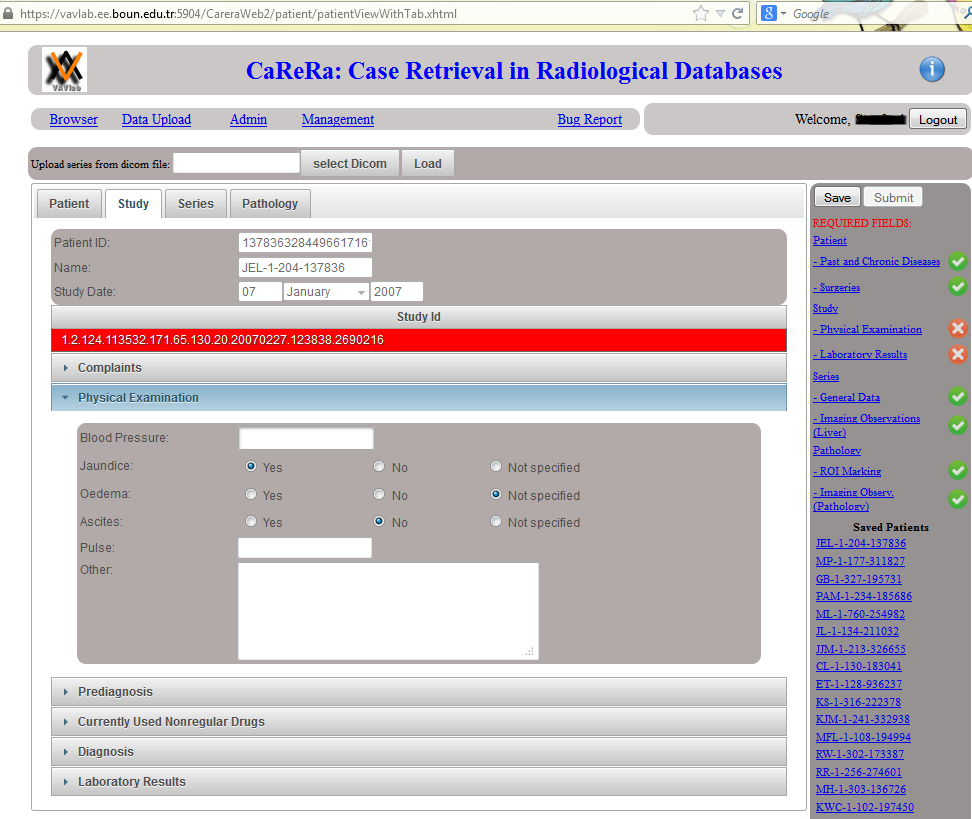
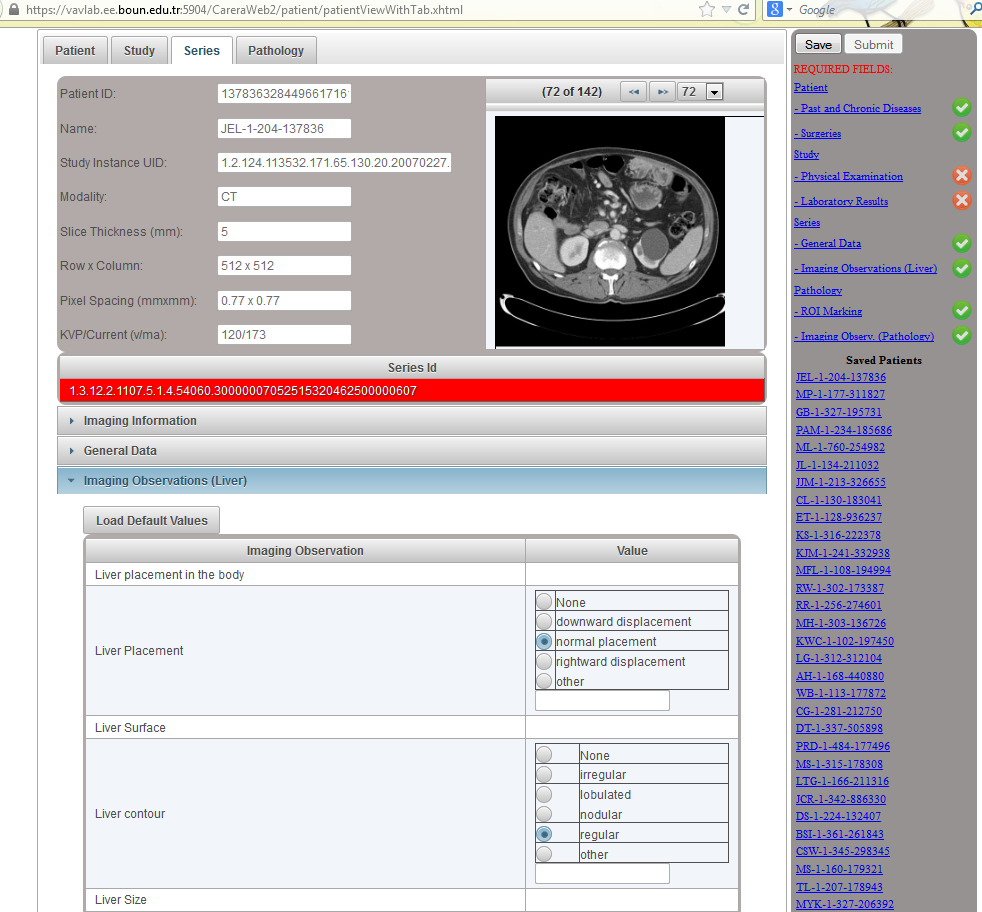
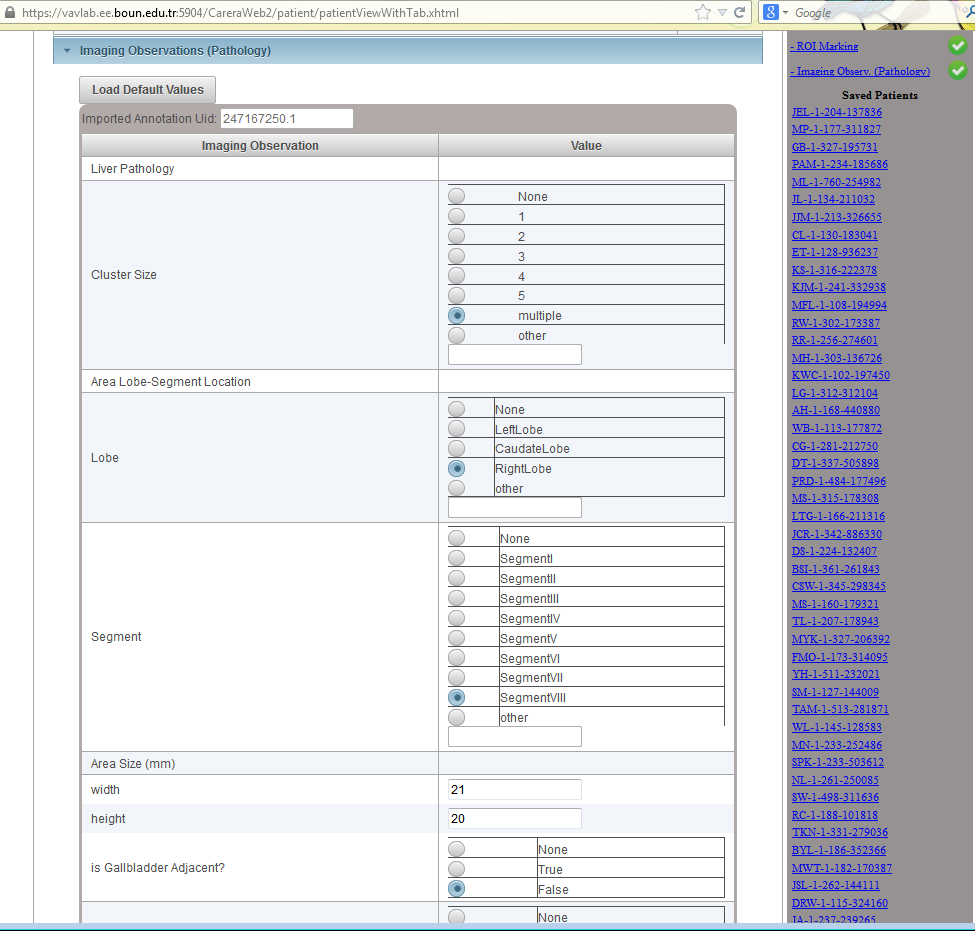
Data Upload: On the right hand side, a list of required fields are displayed. Whenever a required field is filled with some data, a green check mark (otherwise a red cross mark) is shown. Once the radiologist provides information about all required fields, the patient can be submitted. Otherwise, the patient information is saved and can be completed later.
Patient Tab: In this tab, patient information (demographic, regular drugs, past and chronic diseases, surgeries and genetic diseases) is provided. Past and chronic diseases of a patient are listed. Note that diseases are provided using ICD-10 codes. Other diseases are listed as free text.
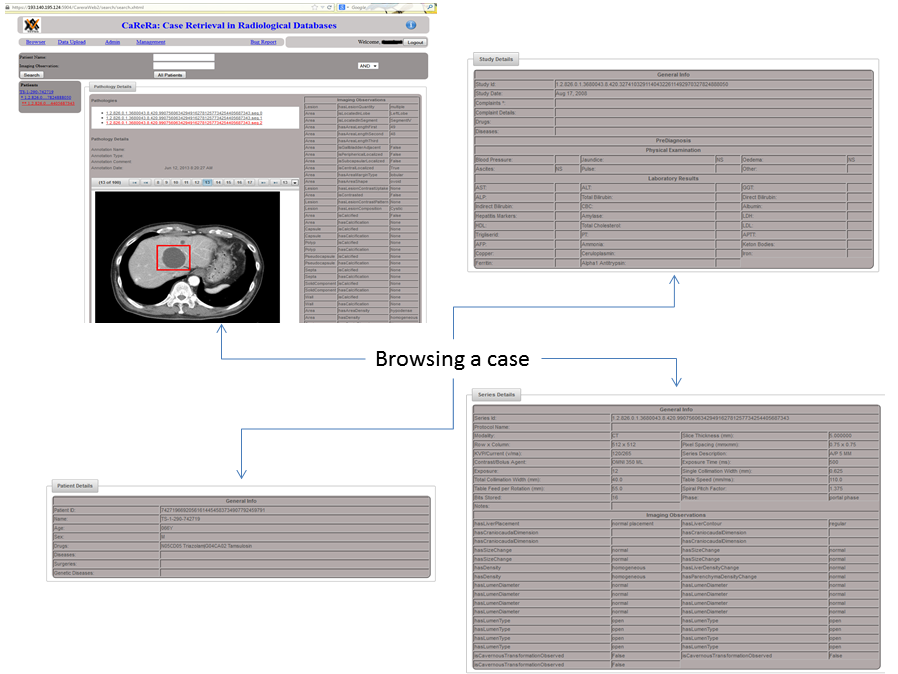


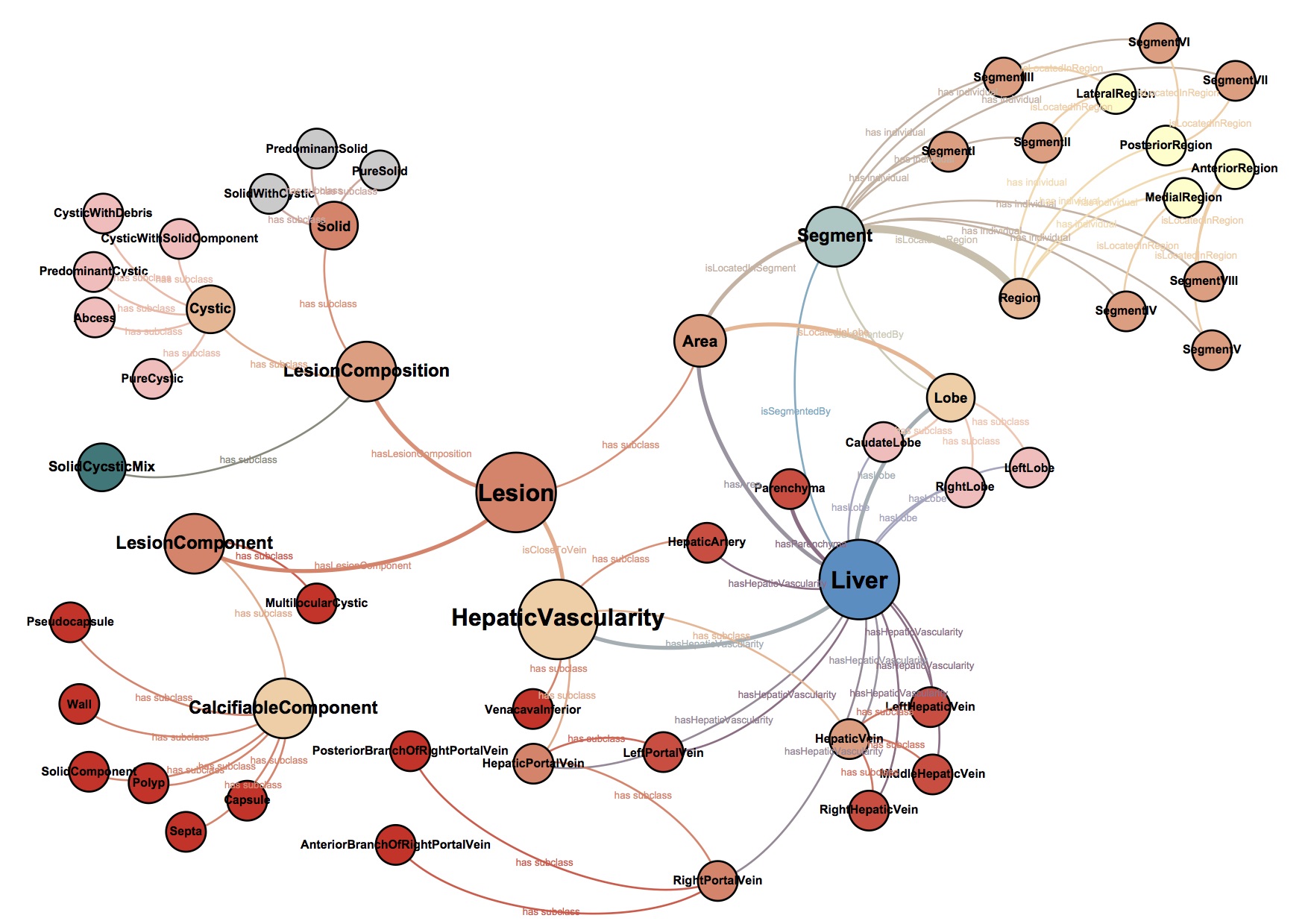
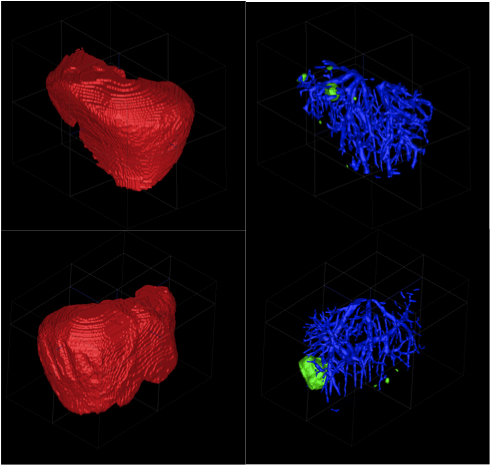
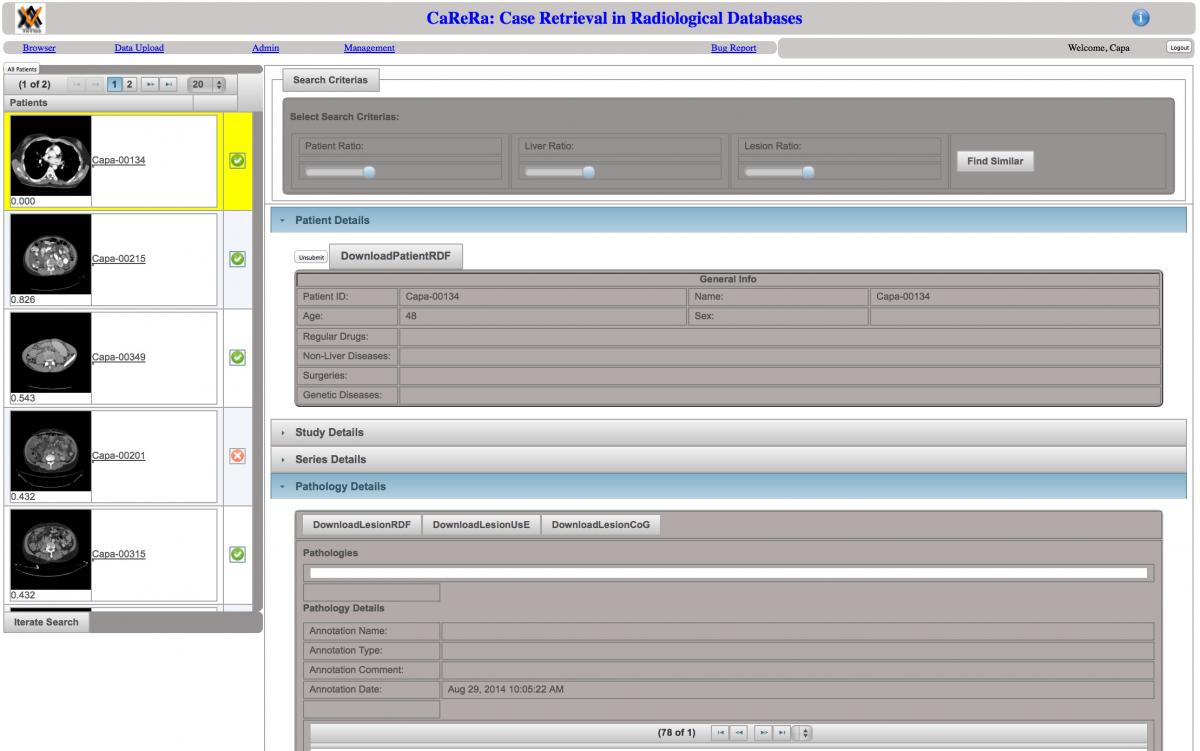
 Based on the fact that clinical experience plays a key role in the performance of medical professionals, it is conjectured that a Clinical Experience Sharing (CES) platform, i.e. a searchable collective clinical experience knowledge-base accessible by a large community of medical professionals, would be of great practical value in clinical practice as well as in medical education. Such a CES would be composed of a multi-modal medical case database, would incorporate a Content Based Case Retrieval (CBCR) engine and would be specialized for different domains. Project CaReRa aims at developing such a CES for the domain of liver cases. During the course of the project, multi-modal case data will be collected, anonymized and stored in a structural database, CBCR technologies will be developed, experiments for the assessment of its impact on the clinical workflow as well as medical education will be designed and conducted.
Based on the fact that clinical experience plays a key role in the performance of medical professionals, it is conjectured that a Clinical Experience Sharing (CES) platform, i.e. a searchable collective clinical experience knowledge-base accessible by a large community of medical professionals, would be of great practical value in clinical practice as well as in medical education. Such a CES would be composed of a multi-modal medical case database, would incorporate a Content Based Case Retrieval (CBCR) engine and would be specialized for different domains. Project CaReRa aims at developing such a CES for the domain of liver cases. During the course of the project, multi-modal case data will be collected, anonymized and stored in a structural database, CBCR technologies will be developed, experiments for the assessment of its impact on the clinical workflow as well as medical education will be designed and conducted.



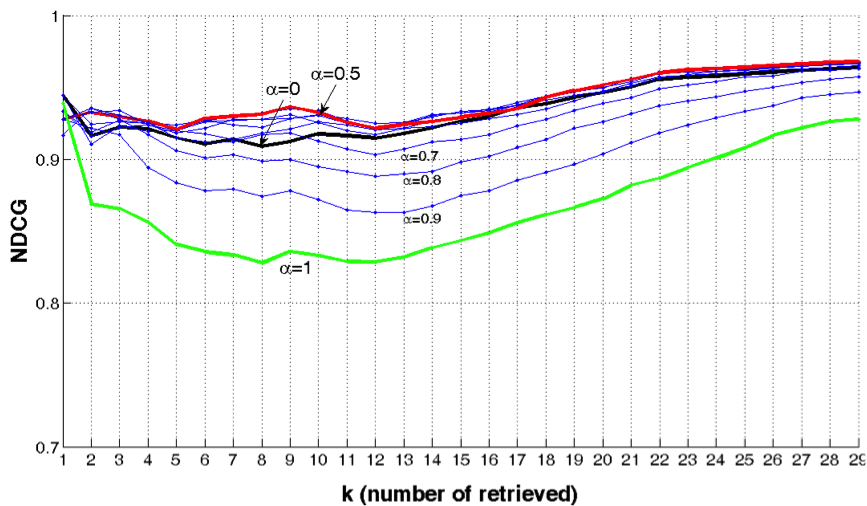
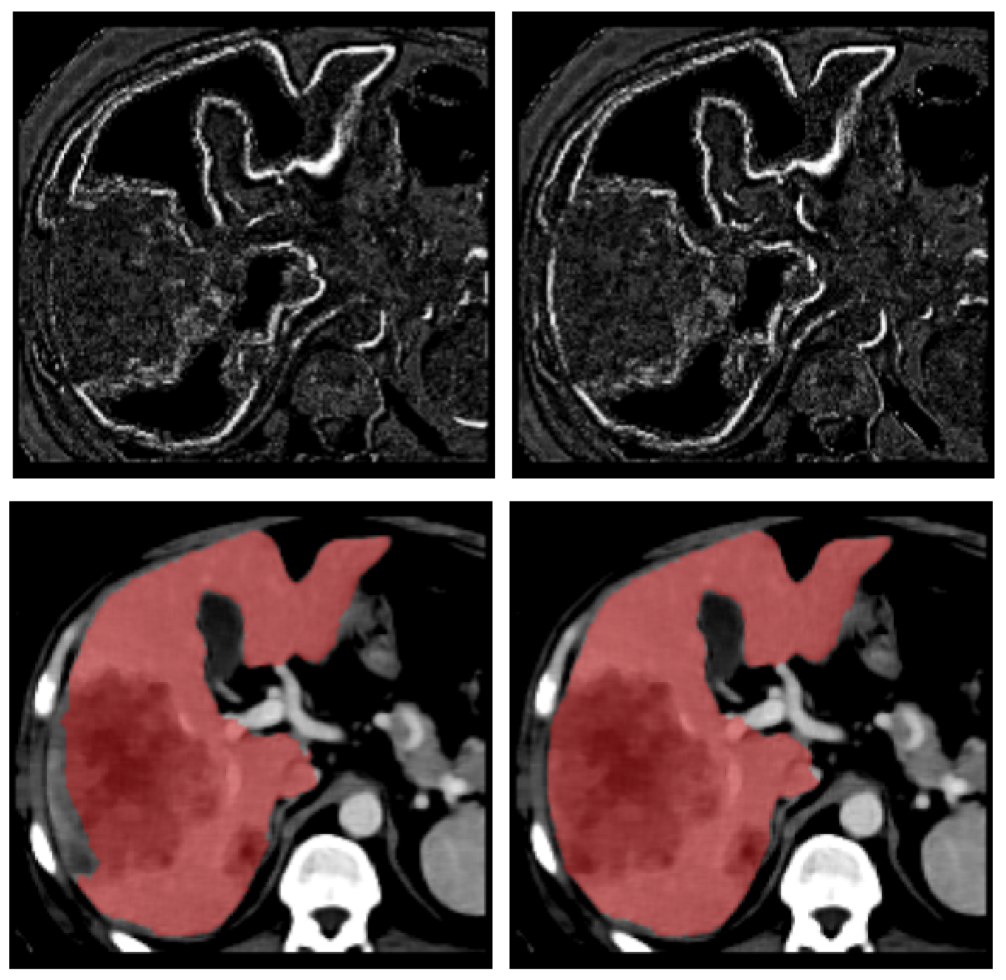
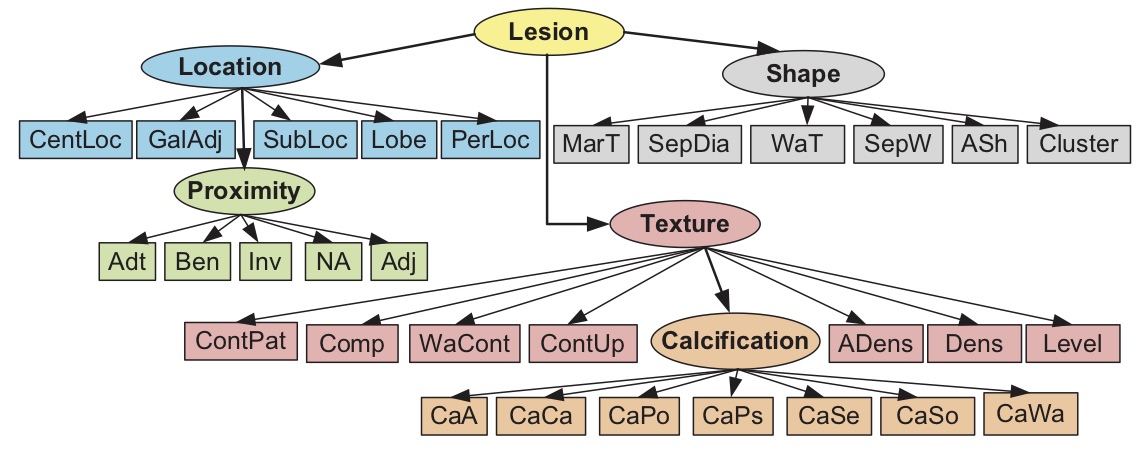 A radiologist-in-the- loop semi-automatic CMIA system is proposed. It is based on a Bayesian tree structured model, linked to RadLex. The experiments with liver lesions in computed tomography (CT) images. show that on average 7.50 (out of 29) manual annotations is sufficient for 95% accuracy in liver lesion annotations. The proposed system guides the radiologist to input the most critical information in each iteration and uses a network model to update the full annotation online. The results also suggest that the domain-aware models perform better than the domain-blind models learned from data.
A radiologist-in-the- loop semi-automatic CMIA system is proposed. It is based on a Bayesian tree structured model, linked to RadLex. The experiments with liver lesions in computed tomography (CT) images. show that on average 7.50 (out of 29) manual annotations is sufficient for 95% accuracy in liver lesion annotations. The proposed system guides the radiologist to input the most critical information in each iteration and uses a network model to update the full annotation online. The results also suggest that the domain-aware models perform better than the domain-blind models learned from data.