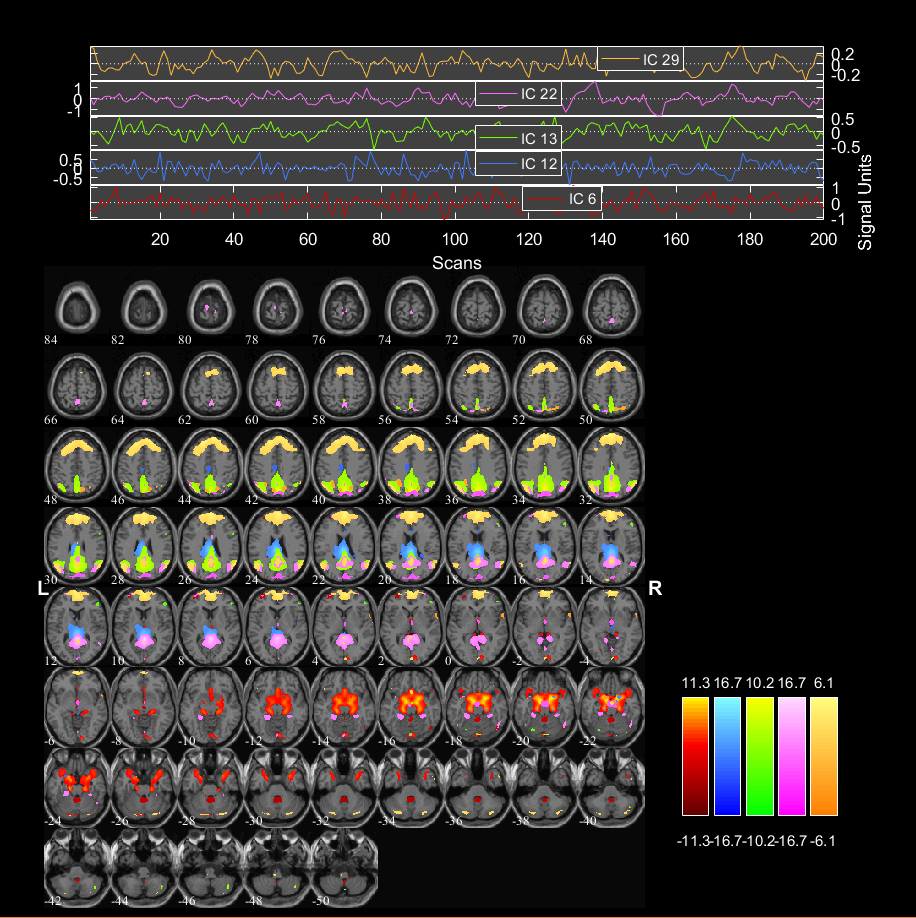
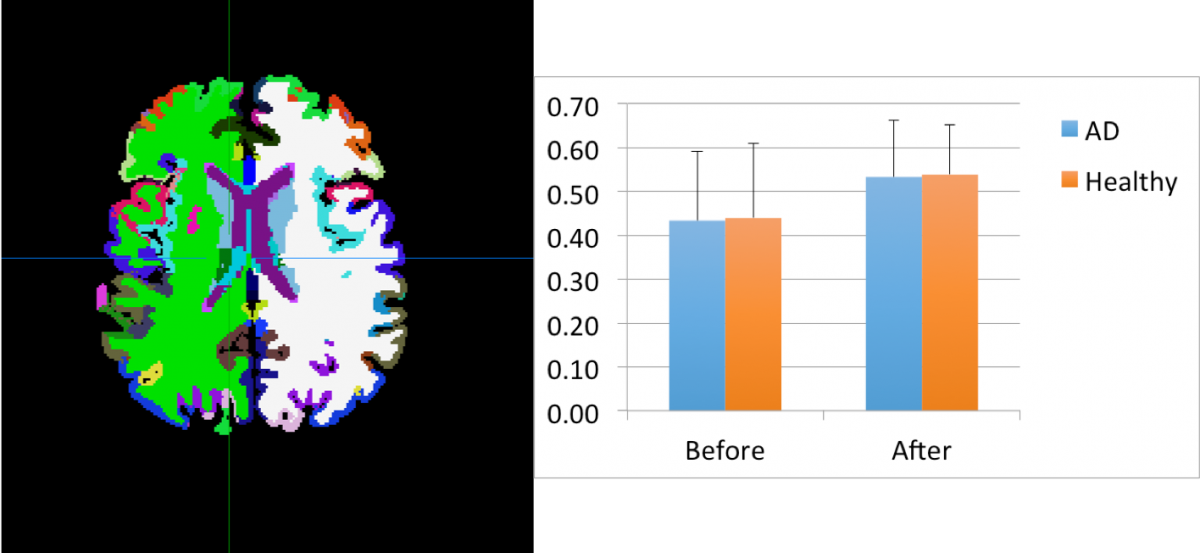
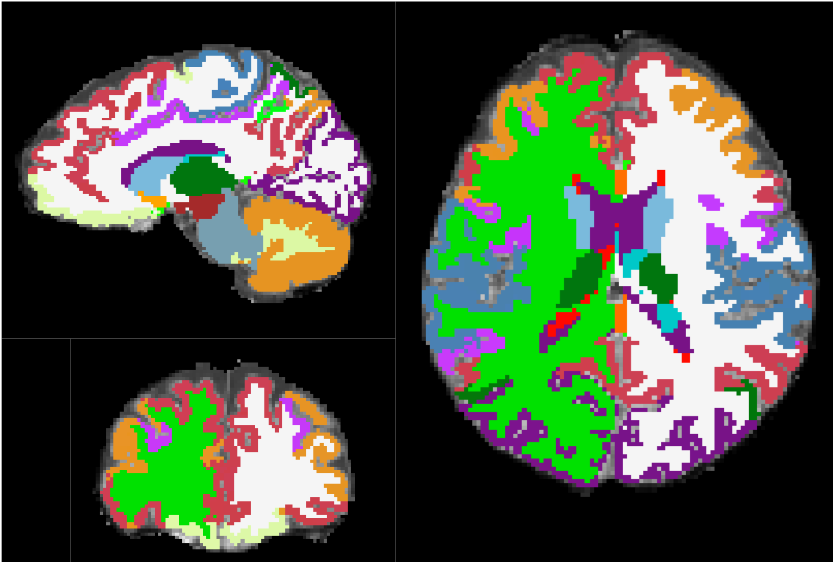
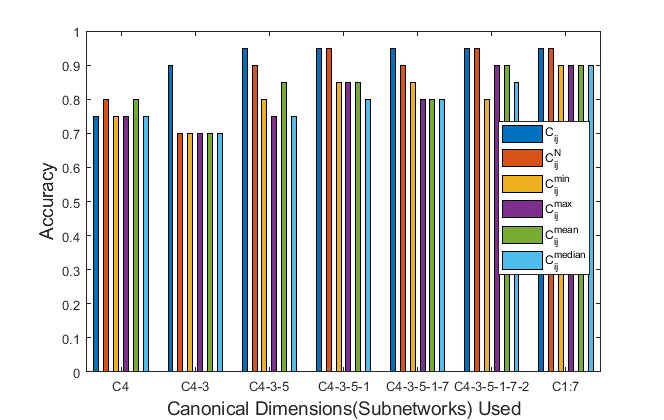
Diffusion MRI & Tractography:
- Le Bihan, D, et al., Artifacts and pitfalls in diffusion MRI, JOURNAL OF MAGNETIC RESONANCE IMAGING (2006)
- Basser, PJ, et al, Diffusion-tensor MRI: theory, experimental design and data analysis - a technical review, NMR IN BIOMEDICINE (2002)
- Wu, WC, et.al., Image Formation in Diffusion MRI: A Review of Recent Technical Developments, JOURNAL OF MAGNETIC RESONANCE IMAGING (2017)
- Alexander, DC, et al., Imaging brain microstructure with diffusion MRI: practicality and applications, NMR IN BIOMEDICINE (2019)
- Jeurissen, B., et al., Diffusion MRI fiber tractography of the brain, NMR IN BIOMEDICINE (2019)
- Mori, S., et al., Fiber tracking: principles and strategies - a technical review, NMR IN BIOMEDICINE (2002)
- Jenkinson, M., et al., FSL, NEUROIMAGE (2012)
- Tournier, JD, et.al., MRtrix3: A fast, flexible and open software framework for medical image processing and visualisation, NEUROIMAGE (2019)
Brain Connectomes:
- Sporns O, Structure and function of complex brain networks, Dialogues Clin. Neuroscience (2013 Sep 1)
- Bullmore E, Sporns O, Complex brain networks: graph theoretical analysis of structural and functional systems, Nat Rev Neurosci (2009 Mar 1) 10: 186-98
- Fornito A, et al, Graph analysis of the human connectome: promise, progress, and pitfalls, Neuroimage (2013 Oct 15) 80: 426-44
- Kaiser Marcus, A tutorial in connectome analysis: Topological and spatial features of brain networks. NeuroImage (2011 Jan 1) 57: 892-907
- Zhang, F., et al., Quantitative mapping of the brain's structural connectivity using diffusion MRI tractography: A review, NEUROIMAGE (2022)
- Zalesky A, Fornito A, Bullmore ET. Network-based statistic: identifying differences in brain networks. NeuroImage (2010) 53:1197-207
- Hagmann P, et al, Mapping the Structural Core of Human Cerebral Cortex. PLoS Biology (2008 Jan 1) 6: e159
- Bastiani M, et al, Human cortical connectome reconstruction from diffusion weighted MRI: The effect of tractography algorithm. Neuroimage (2012 Jun 12) 62: 1732-1749
- Smith SM, et al, Network modelling methods for FMRI. Neuroimage (2011 Jan 15) 54: 875-91
- Horn A, Blankenburg F. Toward a standardized structural-functional group connectome in MNI space. Neuroimage (2016 Jan 1) 124 (Pt A): 310-322.
- Zhu Dajiang, et al, Fusing DTI and FMRI Data: A Survey of Methods and Applications. NeuroImage (2013 Jan 1)
- Griffa A, Baumann PS, Thiran JP, Hagmann P. Structural connectomics in brain diseases. Neuroimage (2013 Oct 15) 80: 515-26
- Cichocki A.Tensor Decompositions: A New Concept in Brain Network Analysis? (2013) http://arxiv.org/abs/1305.0395
-
Zhang Z, Allen GI, Zhu H, Dunson D. Tensor network factorizations: Relationships between brain structural connectomes and traits. Neuroimage (2019 Aug 15) 197: 330-343
- Preti M, Van De Ville D. Decoupling of brain function from structure reveals regional behavioral specialization in humans. Nature Communications, (2019), 1-7, 10(1)
- Graph Signal Processing, Schuman et al, 2013
- Representation Learning on Graphs, Hamilton et al. 2017
- Stanford Graph Learning Workshop, 2022
- Graph Neural Networks (GNNs)





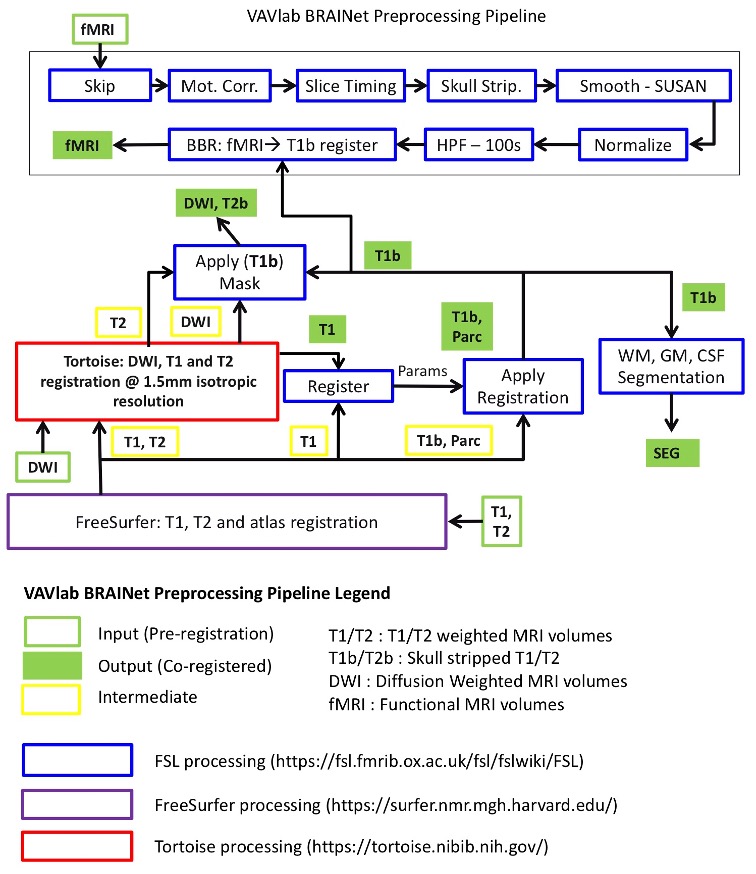
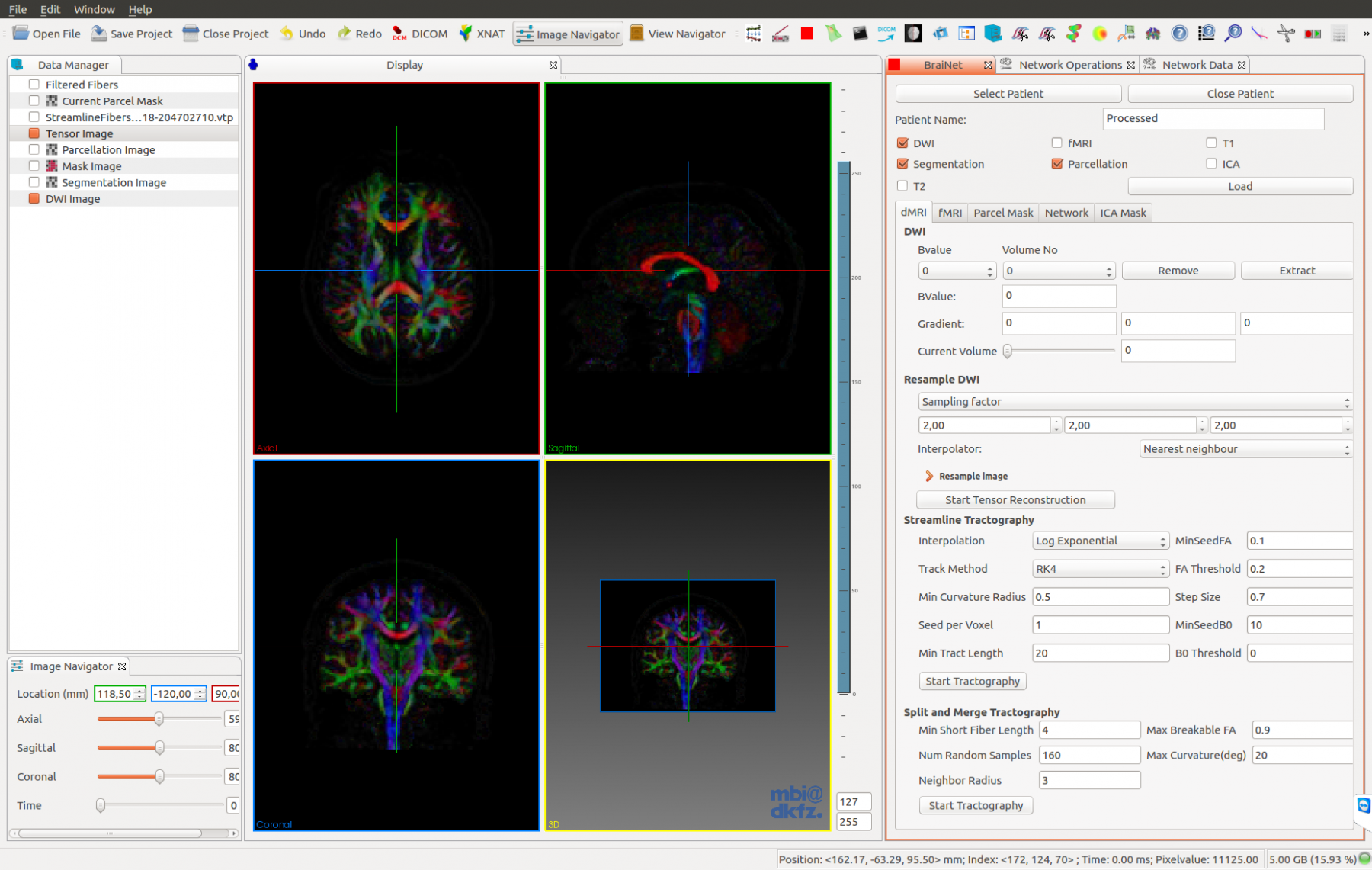
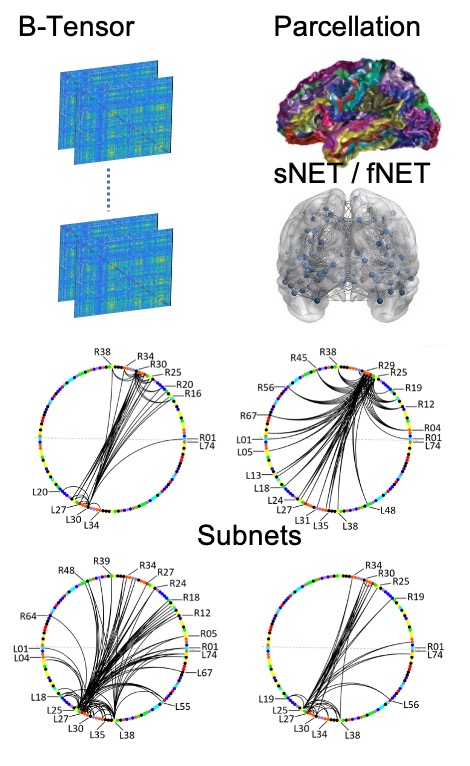 A promising approach in AD research is the analysis of structural and functional brain connectomes, i.e. sNETs and fNETs, respectively. We propose to use tensor representation (B-tensor) of uni-modal and multi-modal brain connectomes to define a low-dimensional space via tensor factorization. The proposed B-tensor encapsulates the whole population either with
A promising approach in AD research is the analysis of structural and functional brain connectomes, i.e. sNETs and fNETs, respectively. We propose to use tensor representation (B-tensor) of uni-modal and multi-modal brain connectomes to define a low-dimensional space via tensor factorization. The proposed B-tensor encapsulates the whole population either with